SPIND Manual
SPIND can work in two modes: CrystFEL-plugin or stand-alone mode, to extract peaks, index pattern and merge intensity.
CrystFEL-plugin mode
For most SFX(Serial Femtosecond Crystallography) users, especially people who are familiar with CrystFEL, plugin mode is recommended. Users should install the modified CrystFEL with SPIND module(crystfel_spind.tar). If not, please see here for installation.
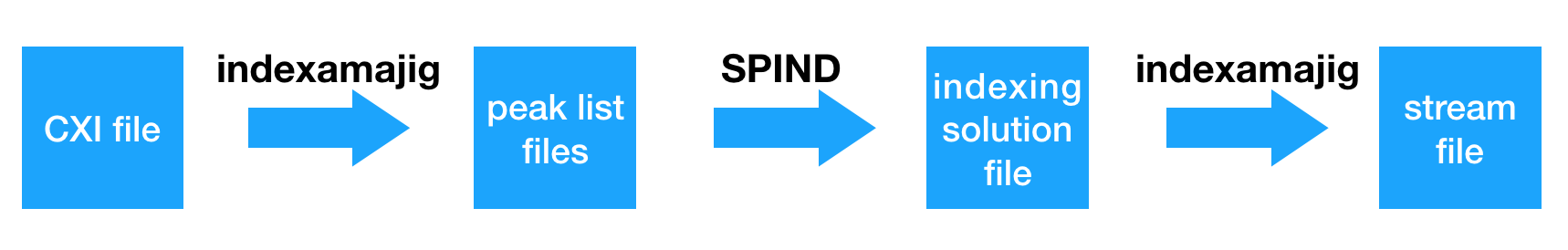
Extract Peaks
indexamajig -i events.lst -o extract_peaks.stream -g detector.geom --peaks=cxi --indexing=spind --felix-options="spind_dir=spind_directory,extract_peaks"
peak_directory is absolute path of work directory for SPIND.
Index with SPIND
There are two steps to index with SPIND.
First, use generate_table.py scripy to generate reference table.
python generate_table.py config.yml
Then perform indexing using SPIND.py script.
python SPIND.py config.yml
All options and parameters are specified in a configuration file.
# configuration example
# lattice information
cell parameters: [103.45,50.28,69.380,90.00,109.67,90.00] # in A and degree
# exp. parameters
wave length : 1.306098E-10 # in meters
detector distance : 136.4028E-3 # in meters
pixel size : 110.E-6 # in meters
# reference generation parameters
resolution cutoff: 4.5E-10 # highest resolution for reference, in meters
lattice type: "monoclinic" # support monoclinic, orthorhombic
centering: "C" # centering type, optional, support I, A, B, C, P
reference table: "spind_reference.h5" # table filename
# indexing parameters
peak list directory: "peak_list"
output directory: "output"
sort by: "snr" # peak list sort method, support snr, intensity
seed pool size: 5 # generate seed from this pool. 5 is good for SFX
refine cycles: 10 # refine indexing solution
seed length tolerance: 3.E+7 # in m^-1, depends on sample and exp. setup
seed angle tolerance: 1.0 # in degrees
seed hkl tolerance: 0.1 # seed hkl tolerence
centering factor: 0.2 # centering weighting factor
eval tolerance: 0.25 # pair peak criterion
multi index: False # try to index multiple crystal for single pattern?
# first event: 0 # first event for indeixng, optional
# last event: 100 # last event for indexing, optional
hkl file: None # txt file including all possiable miller indices, optional
hkl constraint: False # if apply strict hkl constraint, users must specify a hkl file.
Post-processing with CrystFEL
First we need convert SPIND indexing solution into stream files.
indexamajig -i events.lst -o spind.stream -g detector.geom --peaks=cxi --indexing=spind --felix-options="spind_dir=peaks_directory" -p prot.cell
Users can also add other options of indexamajig, see details here
When the stream files ready, users can use CrystFEL programs for visualization, merging…
Authors & Contributors:
- Chufeng Li(Proposer) chufengl@asu.edu
- Xuanxuan Li(Developer) lxx2011011580@gmail.com
- Richard Kirian rkirian@asu.edu
- Nadia Zatsepin Nadia.Zatsepin@asu.edu
- John Spence spence@asu.edu
- Haiguang Liu hgliu@csrc.ac.cn